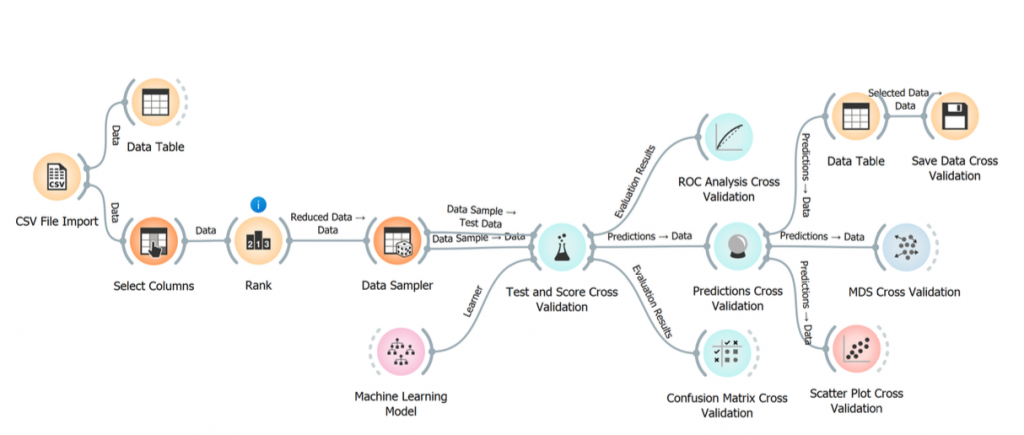
Have a look at our recent collaborative research paper published in Journal of Computer-Aided Molecular Design (IF 4.179), a collaboration with João Carneiro (CIIMAR), Manuel Simões (LEPABE, FEUP) and Diogo Pratas (Univ Aveiro). In this paper we have developed a framework for the identification of the protein targets of molecules with experimentally confirmed antibiofilm activity using machine-learning.
This framework, called TargIDe is based on an ensemble machine learning classification model following a a selection of nine ML models following initial tests involving the development of 27 different ML models using KNN, SVM, NN, NBC, RF and XGBoost.
TargIDe: a machine-learning workflow for target identification of molecules with antibiofilm activity against Pseudomonas aeruginosa
João Carneiro, Rita P. Magalhães, Victor M. de la Oliva Roque, Manuel Simões, Diogo Pratas and Sérgio F. Sousa
Journal of Computer-Aided Molecular Design (2023) | DOI: 10.3390/ph16040632
Abstract
Bacterial biofilms are a source of infectious human diseases and are heavily linked to antibiotic resistance. Pseudomonas aeruginosa is a multidrug-resistant bacterium widely present and implicated in several hospital-acquired infections. Over the last years, the development of new drugs able to inhibit Pseudomonas aeruginosa by interfering with its ability to form biofilms has become a promising strategy in drug discovery. Identifying molecules able to interfere with biofilm formation is difficult, but further developing these molecules by rationally improving their activity is particularly challenging, as it requires knowledge of the specific protein target that is inhibited. This work describes the development of a machine learning multitechnique consensus workflow to predict the protein targets of molecules with confirmed inhibitory activity against biofilm formation by Pseudomonas aeruginosa. It uses a specialized database containing all the known targets implicated in biofilm formation by Pseudomonas aeruginosa. The experimentally confirmed inhibitors available on ChEMBL, together with chemical descriptors, were used as the input features for a combination of nine different classification models, yielding a consensus method to predict the most likely target of a ligand. The implemented algorithm is freely available at https://github.com/BioSIM-Research-Group/TargIDe under licence GNU General Public Licence (GPL) version 3 and can easily be improved as more data become available.