Very happy to announce our recent collaborative paper on the description of Non-B DNA conformations through molecular dynamics simulations, a collaboration of BioSIM with João Carneiro (CIIMAR) and Luisa Azevedo (I3S), published in Biochimica et Biophysica Acta (BBA) – General Subjects (IF: 4.117)
In this paper, we present and optimize a multi-level methodology for the prediction and characterization of tridimensional non-B DNA conformations. The fine-scale atomic molecular determination of this type of non-B conformation opens the way to perform computational molecular studies that can show their involvement in mtDNA cellular mechanisms.
Details can be found in our article.
Non-B DNA Conformations Analysis Through Molecular Dynamics Simulations
André F. Pina, Sérgio F. Sousa, Luísa Azevedo, João Carneiro
BBA – General Subjects | DOI: 10.1016/j.bbagen.2022.130252
Abstract
Background
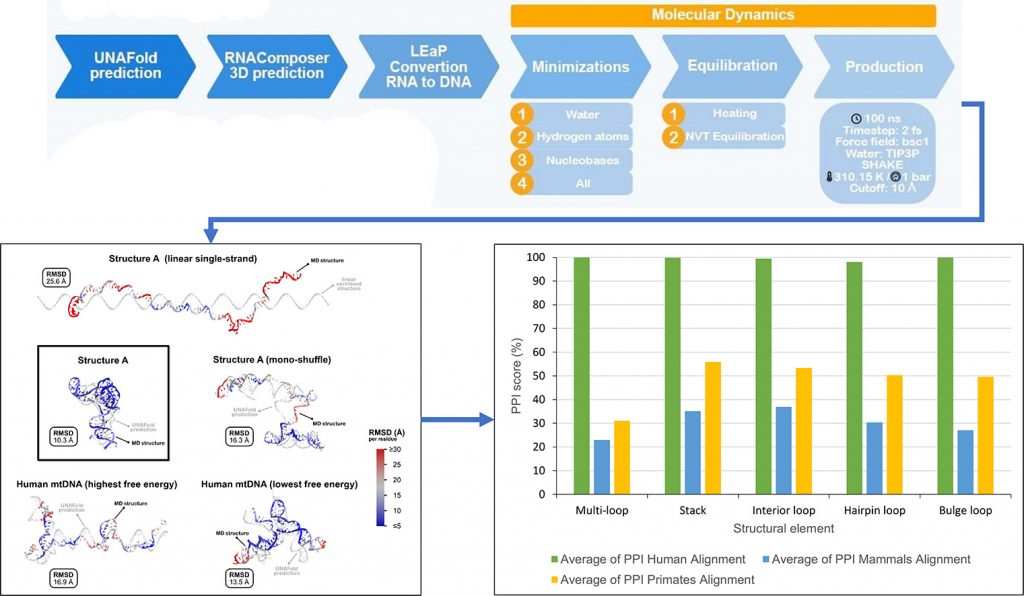
Non-B DNA conformations are molecular structures that do not follow the canonica DNA double helix. Mutagenetic instability in nuclear and mitochondrial DNA (mtDNA) genomes has been associated with simple non-B DNA conformations, as hairpins or more complex structures, as G-quadruplexes. One of these structures is Structure A, a cloverleaf-like non-B conformation predicted for a 93-nt (nucleotide) stretch of the mtDNA control region 5′-peripheral domain. Structure A is embedded in a hot spot for the 3′ end of human mtDNA deletions revealing its importance in influencing the mutational instability of the mtDNA genome.
Methods
To better characterize Structure A, we predicted its 3D conformation using state-of-art methods and algorithms. The methodologic workflow consisted in the prediction of non-B conformations using molecular dynamics simulations. The conservation scores of alignments of the Structure A region in humans, primates, and mammals, was also calculated.
Results
Our results show that these computational methods are able to measure the stability of non-B conformations by using the level of base pairing during molecular dynamics. Structure A showed high stability and low flexibility correlated with high conservation scores in mammalian, more specifically in primate lineages.
Conclusions
We showed that 3D non-B conformations can be predicted and characterized by our methodology. This allowed the in-depth analysis of the structure A, and the main results showed the structure remains stable during the simulations.
General significance
The fine-scale atomic molecular determination of this type of non-B conformation opens the way to perform computational molecular studies that can show their involvement in mtDNA cellular mechanisms.