Very proud to finally announce our most recent research article on SARS-CoV-2, a multidisciplinary collaboration of BioSIM involving University of Bordeaux, CNRS, Nantes Université, Université de Paris, ENS Paris Saclay, Université d’Orléans, showing the power of computational tools, including docking and virtual screening, in drug discovery.
This study, which started in March2020 during the first SARS-CoV-2 lockdown promoted by the Crowdfight COVID initiative, describes the identification and experimental validation of 2 new inhibitors of SARS-CoV-2 cellular entry. The compounds reported here constitute the first molecules that act on the SARS-CoV-2 entry step by specifically targeting the interaction of the spike protein of SARS-CoV-2 and the human ACE2 receptors, which makes them very promising agents. Patents covering these two drugs have already been submitted on 2021.
Computers are amazing! This study involved the computational evaluation of ca. 140 000 molecules with atomic level detail at BioSIM using our in-house designed protocols and our 3D models of the SARS-CoV-2 spike protein (S) and of the interaction with the human ACE2 receptor. The computational modeling studies led to a selection of the 10 most promising molecules for experimental evaluation by an international multidisciplinary research team by both cellular and biochemical approaches, to assess their impact on SARS-CoV-2 entry and on S/ACE2 interaction.
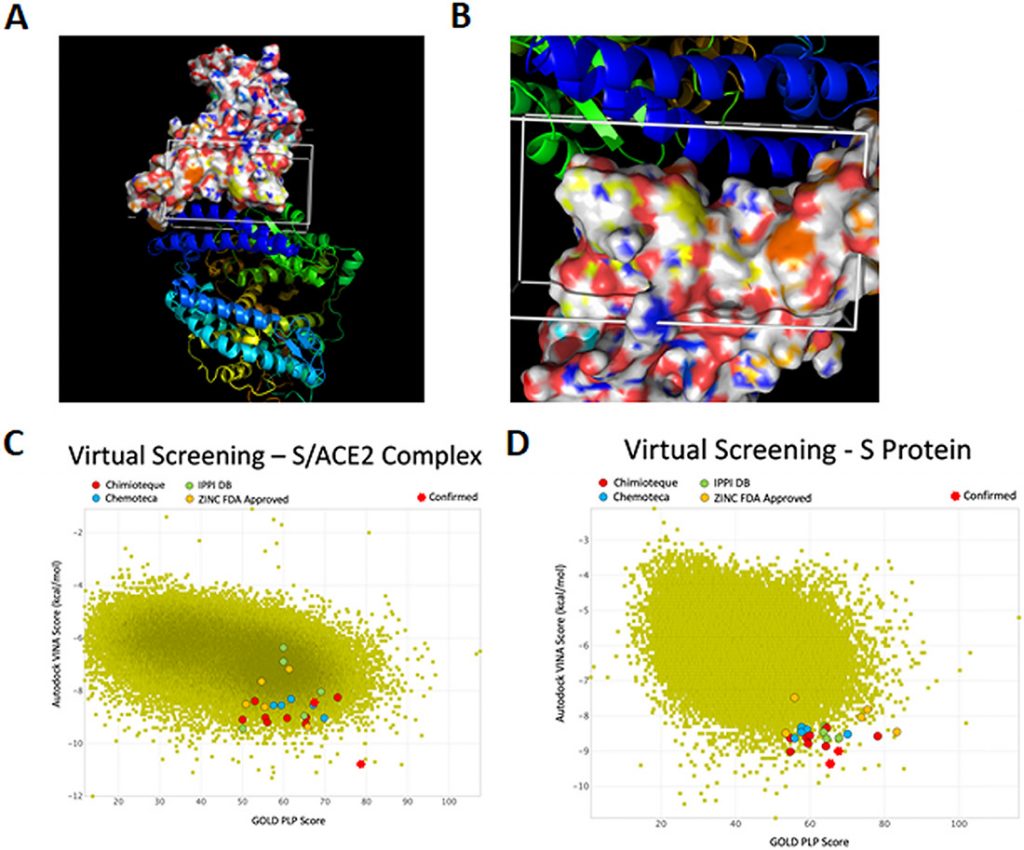
Selection of drugs targeting the S/ACE2 interaction by virtual screening studies. (A and B) The virtual screenings targeting the reported region in the S/ACE2 complex and in S alone were performed by molecular dynamics calculations. The surface representation corresponds to S, while the ribbon representations represent ACE2. (C and D) Results of the virtual drug screening.
2 of the 10 molecules selected from the computational studies were shown experimentally to effectively block the infectivity of lentiviral vectors pseudotyped with the SARS-CoV-2 S protein as well as wild-type and the circulating variant SARS-CoV-2 strains in various human cell lines, including pulmonary cells naturally susceptible to infection. In addition, AlphaLISA and biolayer interferometry confirmed a direct inhibitory effect of these two drugs on the S/ACE2 association. These molecules were shown to inhibits viral replication with a 50% effective concentration (EC50) between 0.1 and 0.5 μM depending on the cell lines.
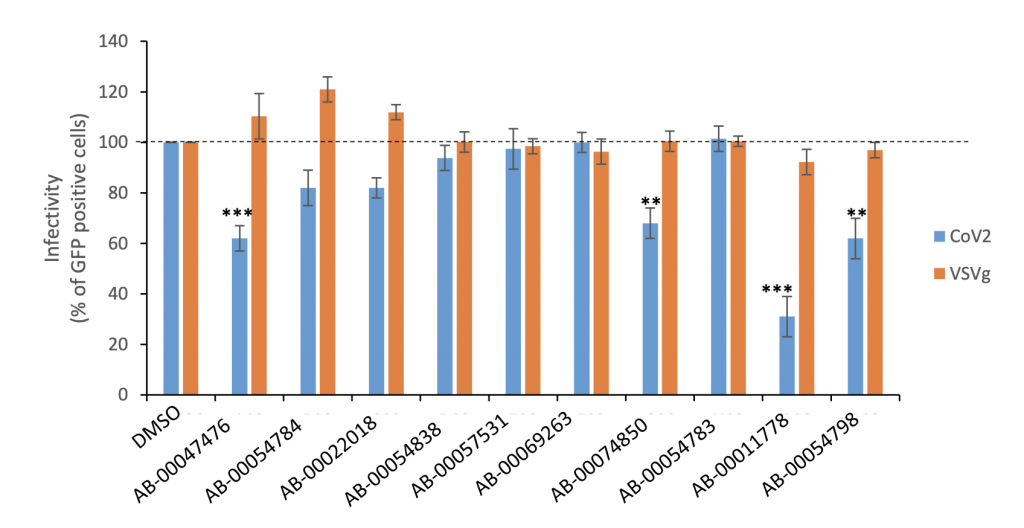
Experimental screening of the 10 drugs selected from the computational studies on inhibiting SARS-CoV-2 entry using a lentiviral pseudovirus system. Drugs were tested for their infectivity of both VSVg and SARS-CoV-2 S pseudotyped lentiviruses in HEK293T-ACE2 cells at 10μM. 4 out of the 10 molecules selected from the computational studies inhibited SARS-CoV-2 entry. AB-00047476 and AB-00011778 were selected for further characterization using increasing concentrations of drug. Data are shown as the means of at least three independent experiments ± the standard deviations. ***p<0.001, **p<0.01 (Student’s t-test).
Aside from the recently licensed remdesivir and nirmatrelvir-ritonavir (Paxlovid), no effective drugs have been validated as potential specific anti-COVID treatments. Thus, the compounds reported here constitute the first molecules that act on the SARS-CoV-2 entry step by specifically targeting the S/ACE2 interfaces, making them very promising agents. A comparison of AB-00011778, remdesivir, and ritonavir (one of the two antiproteases included in Paxlovid) showed that while the remdesivir remains the most efficient molecule known to date, our molecule AB-00011778 was more efficient than the ritonavir molecule in inhibiting the SARS-CoV-2 replication in human pulmonary A549-ACE2 cells.
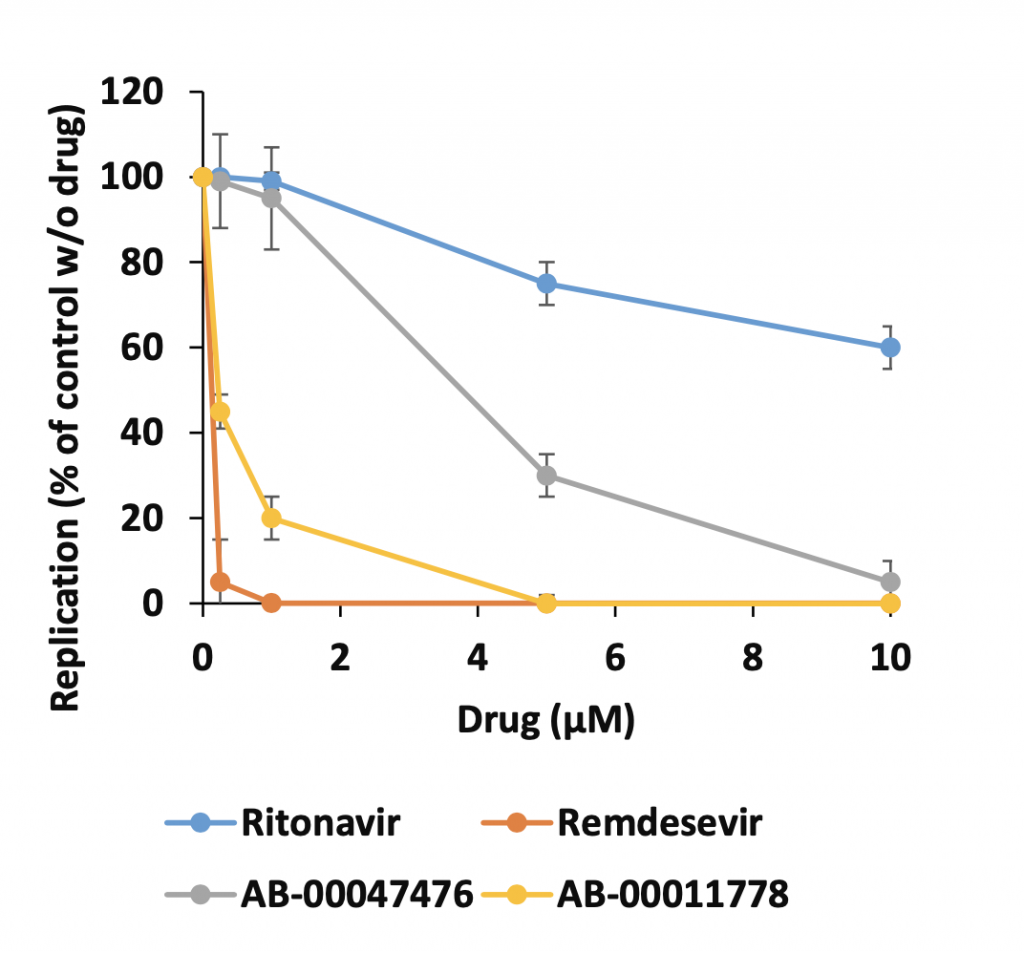
Comparison of the effects of Remdesivir, Ritonavir, AB-00011778 and AB-00047476 on SARS-CoV-2 replication in human pulmonary cells. A549-ACE2 were incubated with the SARS-CoV-2 reference strain (MOI=1) and increasing concentrations of the tested compounds. Replication was evaluated by quantifying the viral genome copies after 24 hours. Data are reported as the means from two independent experiments and are expressed as a percentage of the control without molecule ± the standard deviations.
Of note, in contrast to the repurposed remdesivir and ritonavir drugs acting on different replication steps, our molecules represent original molecule that have been specifically selected to block SARS-CoV-2 entry and have not yet been optimized. The low cytotoxicity observed in various cell lines also paves the way for future animal model studies and clinical trials to fully evaluate their antiviral properties and therapeutic potency.
More details on this massive study involving 25 researchers and 15 different techniques can be found in our paper.
Selection of Bis-Indolyl Pyridines and Triphenylamines as New Inhibitors of SARS-CoV-2 Cellular Entry by Modulating the Spike Protein/ACE2 Interfaces
Delphine Lapaillerie, Cathy Charlier, Véronique Guyonnet-Dupérat, Emilie Murigneux, Henrique S. Fernandes, Fábio G. Martins, Rita P. Magalhães, Tatiana F. Vieira, Clémence Richetta, Frédéric Subra, Samuel Lebourgeois, Charlotte Charpentier, Diane Descamps, Benoît Visseaux, Pierre Weigel, Alexandre Favereaux, Claire Beauvineau, Frédéric Buron, Marie-Paule Teulade-Fichou, Sylvain Routier, Sarah Gallois-Montbrun, Laurent Meertens, Olivier Delelis, Sérgio F. Sousa, Vincent Parissi
Antimicrobial Agents and Chemotherapy | DOI: 10.1128/aac.00083-22
Abstract:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the infectious agent that has caused the current coronavirus disease (COVID) pandemic. Viral infection relies on the viral S (spike) protein/cellular receptor ACE2 interaction. Disrupting this interaction would lead to early blockage of viral replication. To identify chemical tools to further study these functional interfaces, 139,146 compounds from different chemical libraries were screened through an S/ACE2 in silico virtual molecular model. The best compounds were selected for further characterization using both cellular and biochemical approaches, reiterating SARS-CoV-2 entry and the S/ACE2 interaction. We report here two selected hits, bis-indolyl pyridine AB-00011778 and triphenylamine AB-00047476. Both of these compounds can block the infectivity of lentiviral vectors pseudotyped with the SARS-CoV-2 S protein as well as wild-type and circulating variant SARS-CoV-2 strains in various human cell lines, including pulmonary cells naturally susceptible to infection. AlphaLISA and biolayer interferometry confirmed a direct inhibitory effect of these drugs on the S/ACE2 association. A specific study of the AB-00011778 inhibitory properties showed that this drug inhibits viral replication with a 50% effective concentration (EC50) between 0.1 and 0.5 μM depending on the cell lines. Molecular docking calculations of the interaction parameters of the molecules within the S/ACE2 complex from both wild-type and circulating variants of the virus showed that the molecules may target multiple sites within the S/ACE2 interface. Our work indicates that AB-00011778 constitutes a good tool for modulating this interface and a strong lead compound for further therapeutic purposes.

