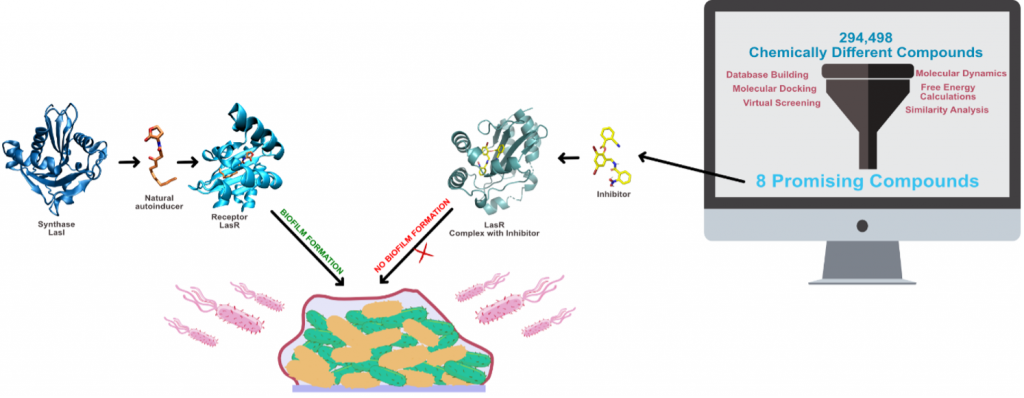
Please check our new research article published in RSC Molecular Systems Design and Engineering (IF: 4.94). In this paper, we have developed and optimized a multi-level in silico virtual screening protocol specifically for LasR, a quorum-sensing receptor of the multidrug-resistant Pseudomonas aeruginosa. We then applied this protocol to screen over 200,000 chemically diverse compounds, to identify 8 novel promising candidates for inhibition of biofilm formation in Pseudomonas aeruginosa.
Identification of novel candidates for inhibition of LasR, a quorum-sensing receptor of multidrug resistant Pseudomonas aeruginosa, through a specialized multi-level in silico approach
Rita P. Magalhães, Tatiana F. Vieira, André Melo and Sérgio F. Sousa
MSDE (2022) | Doi: 10.1039/D2ME00009A
Abstract
The emergence of multi-drug resistant bacteria in the past decades has become one of the major public health issues of our time. One of the main mechanisms of resistance and persistence of bacteria is their ability to form biofilms. Quorum-sensing (QS) is one of the main mechanisms of biofilm formation. Interfering with the QS cascade constitutes a non-antibiotic strategy to reduce biofilm formation and affect bacterial resistance. In Pseudomonas aeruginosa, QS takes place by four different systems, which start with the activation of transcriptional receptor LasR. This receptor is stabilized by C12-HSL. In this work, we have optimized and employed a multi-level in silico protocol to identify promising LasR inhibitors, combining different computer-aided drug design techniques such as molecular docking, virtual screening, molecular dynamics and free energy calculations. The protocol was optimized using all 21 available LasR X-ray structures, 7 docking scoring functions, and a library of 90 active and 4500 inactive compounds. The optimized protocol was used to scan 294 498 chemically distinct compounds from 5 different databases, of which 30 compounds were further studied by molecular dynamics and free energy calculations and resulted in 8 possible QS inhibitors with promising ADME properties and binding affinity.
